Cards In This Set
| Front | Back |
|
What are some diseases caused by picornaviruses?
|
Hepatitismyocardidtispoliofoot-and-mouth disease
|
|
What are some characteristics of picornaviruses?
|
Nonenvelopedicosahedral capsid+ssRNA resitant to pH 3-95' end attached to VPg3' end is polyadenylated
|
|
What four proteins make up the capsid and how many copies of each are there?
|
VP1, VP2, VP3, VP4 60 copies of each
|
|
What makes picornaviruses capsid different than the other viruses?
|
There are no receptors on the capsid surface only depressions called "canyons" or "pits"
|
|
How do Rhinoviruses and Foot-and-mouth disease gain entry to the cell?
|
Via receptor-mediated endocytosis
|
|
How does the poliovirus attach and gain entry into host cell?
|
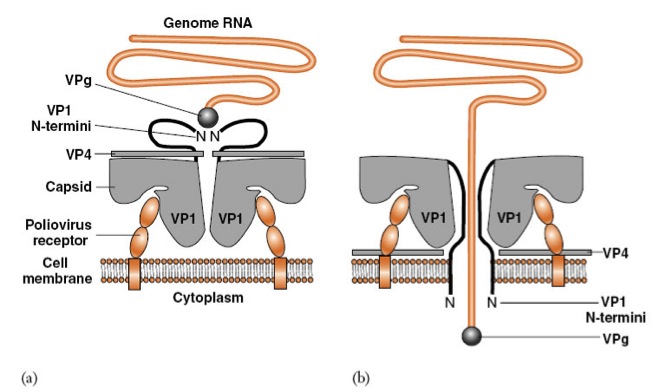 Attachment is via poliovirus receptorVP4, N-term of VP1 and RNA genome (bound to VPg at 5' end) are inside (a) a conformation change causes VP4 to become externalized and the N-term of all 5 VP1 molceules are inserted into the membrane to form a channel for the RNA to pass into the cytoplasm of the host cell |
 Describe cap-dependent initiation of protein synthesis |
MRNA use this to initiate translation near their 5' end eIF-4F complex (eIF-4A, eIF-4E, eIF-4G) recruit a 40S ribosomal subunit to the capped 5' end of mRNA40S will scan mRNA in search of proper AUG codon. 60S ribosomal subunit joins to form an 80S complex and protein synthesis begins
|
|
Is the poly A tract at the 3' end of picornaviruses added?
|
No it is part of the genome sequence
|
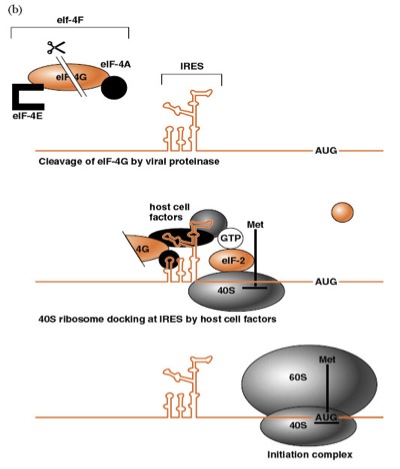 Describe cap-independent initiation of protein synthesis |
Ribosomes are directed to the initiator 5' AUG sequence via an Internal Ribosome Entry Site (IRES) that lies within the highly structured 5' noncoding region. the host cell proteins bind to the IRES and help to dock the 40S onto the RNA. Translocation occurs until initiator AUG is met. the 60S ribosomal subunit will combine to form an 80S and synthesis will occur.
|
|
What are the essential features of IRES?
|
eIF-3,
eIF-4G and eIF-4a promote IRES assembly. A host factor X is also
required.contain a pyrimidine-rich tract 20-25 nt from a conserved AUG codoninitiation of translation is reduced if smaller or larger than 20-25ntGNRA sequence is conserved in specific loop regions of stem-loop structures
|
|
How do picornavirus genome RNAs engineer their translation in host cells?
|
Translation occurs cap-independently.genomes has a 743nt long UTR untraslated leader seq contains 8 AUG codons prior to initiation siteInternal ribosomal binding occurs at the IRES
|
|
What are the IRES ribosomal subunits?
|
ElF-4G (component in eIF4F) cellular initiation factorelF-3Elf-4A- helicase40S ribosomal subunitX (a host factor)
|
|
Key molecules and their specific functions of picornaviruses during translation
|
2A proteinase-cleaves N-term immediately after translation3C Proteinase+precurser 3CD- carry out remaining polyprotein cleavages2B protein-host range determinant involved in RNA synthesis2C protein- highly conserved; involved in RNA synthesis3D- RNA dependend RNA polymerase; has GDD plymerase motif3B = VPg at the 5' end
|
|
Describe viral assembly of picornaviruses
|
1. the P1 precurser polypeptide is cleave by the 3C/3CD proteinase yielding: VP0 VP3 VP1- assembles into 5s protomer2. pentamers of the protomers assemble into a 14S complex (also a pentamer)3a. the 14S complex may then form a 75S empty procapsid OR3b. a 150S complex provirion can be formed (caspid is assembled around viral RNA)4 (only from 3b.) 150S provirion is then made a 160S virion by the clevage of V0--> V2 &V4
|
|
What are some genera in the picornaviridae that don't have an L protein?
|
Enterovirusesrhinoviruses
|



